SpCas9 is an RNA-guided DNA endonuclease and has a facile introduction of genomic alterations. The SpCas9 complex and guide RNA (gRNA) recognizes the substrate sequence via a protospacer adjacent motif (PAM).
Catalytically impaired-SpCas9 to any genomic locus results in transformative technologies. The fusion of catalytically inactive SpCas9 (dCas9) to activators enables gene transcription and repression.
Besides, the fusion of dCas9 to base-modifying enzymes allows base conversion at specific genomic sites. dCas9–GFP fusion can let the imaging of genomic loci possible, Besides, dCas9–acetyltransferases or deacetylases fusion enables epigenome editing.
Several protein anti-CRISPR molecules have been reported. Now, we will introduce small-molecule inhibitors. small-molecule inhibitors are cell-permeable, reversible, and proteolytic stable. Besides, protein-based molecules are always highly potent due to their greater number of SpCas9 interaction sites.
Now, researchers also focus on the study of small-molecule inhibitors. Unlike protein-based anti-CRISPRs, those small molecules always have fast kinetics and inhibit enzymic activity in an extremely quick manner with precise temporal control.
Small molecules have a bright future on its application because of its features.
However, the identification of SpCas9 small-molecule inhibitors exhibits some difficulties. SpCas9 is a single-turnover enzyme and aDNA-binding protein, it holds on to its substrate with picomolar affinity. Besides, the inhibition of SpCas9 activity needs the inactivation of two nuclease domains.
In this article, we will introduce a SpCas9 inhibitor based on a screen of enriched libraries, BRD0539.
BRD0539 is a cell-permeable and non-toxic inhibitor of CRISPR-Cas9. It inhibits Streptococcus pyogenes Cas9 (SpCas9) (apparent IC50=22 μM) in an in vitro DNA cleavage assay。
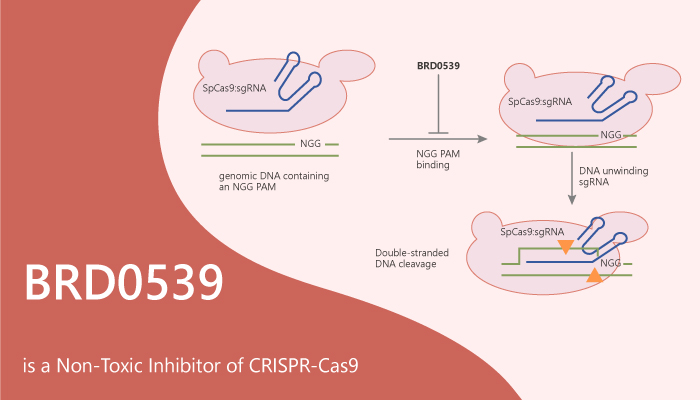
Additionally, BRD0539 dose-dependently blocks the formation of the DNA-bound state in a dose-dependent fashion. It impairs the perturbation induced by the 4PAM DNA, however, it does not interfere with the SpCas9:gRNA interaction.
BRD0539 can inhibit SpCas9 in the eGFP-disruption assay. But it is unable to inhibit FnCpf1, a structurally different CRISPR-associated nuclease, in the same assay, further highlighting the specificity of BRD0539.
In conclusion, as a SpCas9 inhibitor, it lays a foundation for the rapid identification of cell-permeable, reversible, synthetic anti-CRISPR molecules.
Reference:
Basudeb Maji, et al. Cell. 2019 May 2;177(4):1067-1079.e19.